New UK Coronavirus Variant
Rapidly Growing Mutation in the UK With an Unexpectedly Large Number of Genetic Changes
There is now new data from the Journal Cell on the new SarsCov2 variant being seen in southern England based on recent whole-genome sequencing studies.
Viral mutations are like selling real estate…it’s all about location…location…location.
Mutations are essentially typos in the viral genetic code. This most often occurs right after the virus enters a cell and makes copies of its RNA payload through a process called “RNA replication”. Coronaviruses are generally quite resistant to mutations because they carry what’s called a “proofreading enzyme” that can detect and correct for typos.
Most mutations are so-called “synonymous mutations” and are completely benign and don’t alter the meaning or function of the particular stretch of genetic code. But some can create significant functional alterations.
For example, the single-letter typo in “acceptable” vs. “acceptible” is far more acceptable than the single-letter typo in “duck” vs. “fuck”. Same amount of typos…very different results.
What we have with this new SarsCoV2 variant is the “oh duck” sort of typo…not good!
Preliminary Results
The new variant has been labeled VUI-202012/01 and has:
17 functional single-nucleotide variants (8 on the S-gene)
6 variants and 2 deletions in the S-gene (which codes for the spike protein)
6 “synonymous mutations” in ORF1ab and the M-gene (no big deal)
An Orf8 truncation that leaves the virus open to more mutations
71% increase in contagiousness
~2.5x increase in R-value (still very preliminary)
4x higher average viral load
exhibiting exponential growth even during full lockdown (bad!)
currently accounting for 62% of cases in London
no increase in severity of illness (which is a good thing)
So a couple of interesting points. First, this variant has accumulated a huge number of mutations in a very short period of time and this is quite surprising to see.
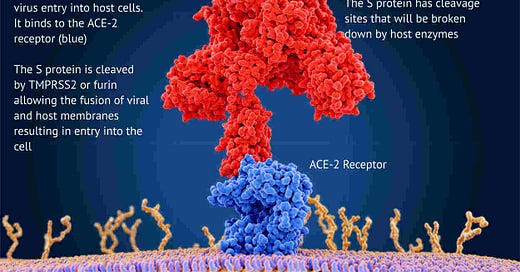
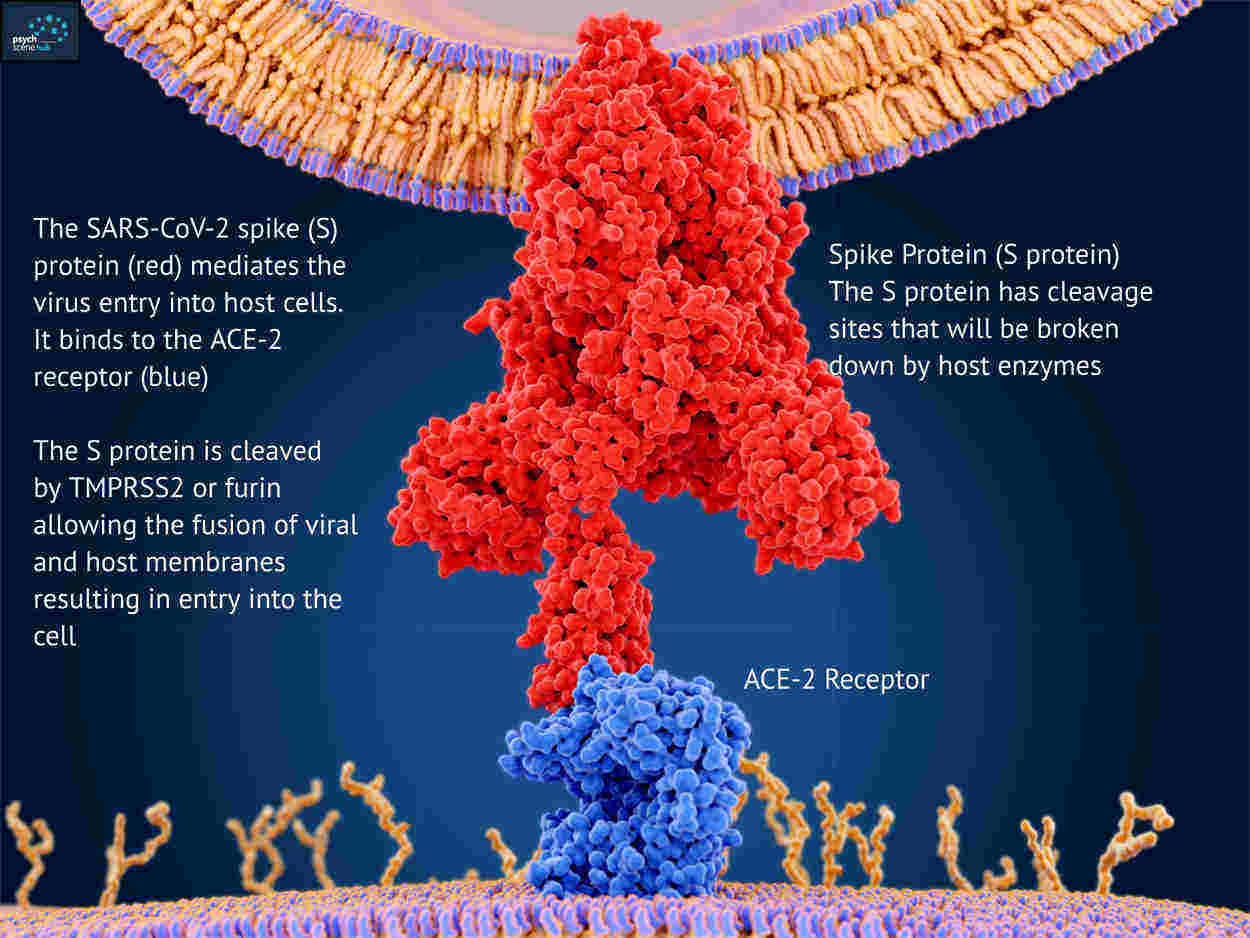
But the most concerning mutations are on the S-gene that carries the instructions for building the spike protein used by the virus to enter cells and used by the vaccines to target the virus.
What Does This Mean?
The virus is rather like a burglar with a lockpick trying to break into your cells. The lockpick is the spike protein that’s used to pick the lock on the front door.
Now let’s say there are a 1000 burglars in your town and suddenly 10 of them got hold of a set of REALLY good lockpicks that makes it a LOT easier to do a break-in. Pretty soon, those 10 burglars would be responsible for almost all the burglaries in town.
This is what’s happening in London right now! These S-gene mutations seem to let the virus break into cells far more efficiently and far quicker than before.
This means a few things:
this variant will be far more contagious
it will infect a person far faster
the viral load will grow faster and to higher levels
However, because the spike protein doesn’t play a role in disease severity, this mutation doesn’t appear to have created a more lethal version of the virus…just a far more contagious version.
Does This Effect the Vaccine?
In short…NO! It’s still a bit too early to know absolutely, but it’s almost certain to have no significant impact on the effectiveness of the vaccine.
The vaccine is designed to look for a large block of amino-acids (about 650+) that make up the head of the spike protein. These mutations result in 2 isolated amino-acid deletions and 4 amino-acid alterations. Such minor changes should have no impact on the ability of the vaccine to find and bind to the correct epitope (the part of the virus the vaccine is targeting).
How Did This Happen?
The sudden appearance of a large group of mutations in an otherwise slow mutating virus is concerning and raises the question of how it might have occurred.
The working hypothesis is that it came from a single infected individual. This person was likely immuno-compromised (due to some unrelated condition) and sustained a long-term severe infection from SarsCoV2. This would provide the right environment for a wide range of mutations to develop.
At some point, the patient was likely treated with Convalescent Plasma and/or Remdisvir. This cleared most of the more normal variants of the virus but left a more highly resistant and more aggressive strain behind that then fully re-infected the patient. And at some point, this variant got spread to other individuals and became a new highly contagious wildtype.
To quote from the research paper:
“What evolutionary processes or selective pressures might have given rise to lineage B.1.1.7?
High rates of mutation accumulation over short time periods have been reported previously in studies of immunodeficient or immunosuppressed patients who are chronically infected with SARS-CoV-2 (Choi et al. 2020; Avanzato et al. 2020; Kemp et al. 2020). These infections exhibit detectable SARS-CoV-2 RNA for 2-4 months or longer (although there are also reports of long infections in some immunocompetent individuals). The patients are treated with convalescent plasma (sometimes more than once) and usually also with the drug remdesivir. Virus genome sequencing of these infections reveals unusually large numbers of nucleotide changes and deletion mutations and often high ratios of non-synonymous to synonymous changes. Convalescent plasma is often given when patient viral loads are high, and Kemp et al. (2020) report that intra-patient virus genetic diversity increased after plasma treatment was given.”
“Under such circumstances, the evolutionary dynamics of and selective pressures upon the intra-patient virus population are expected to be very different to those experienced in typical infection. First, selection from natural immune responses in immune-deficient/suppressed patients will be weak or absent. Second, the selection arising from antibody therapy may be strong due to high antibody concentrations. Third, if antibody therapy is administered after many weeks of chronic infection, the virus population may be unusually large and genetically diverse at the time that antibody-mediated selective pressure is applied, creating suitable circumstances for the rapid fixation of multiple virus genetic changes through direct selection and genetic hitchhiking.”
“These considerations lead us to hypothesise that the unusual genetic divergence of lineage B.1.1.7 may have resulted, at least in part, from virus evolution with a chronically-infected individual. Although such infections are rare, and onward transmission from them presumably even rarer, they are not improbable given the ongoing large number of new infections.”
Reference Links
From the Journal Cell:
Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity
From New Scientist:
What you need to know about the new variant of coronavirus in the UK
From the British Medical Journal:
Covid-19: New coronavirus variant is identified in UK
From the COVID-19 Genomics Consortium UK:
Update on new SARS-CoV-2 variant and how COG-UK tracks emerging mutations
Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations
About the author:
I am a scientist with 20+ years in the biotech industry. I currently work as a consultant with companies involved in developing molecular diagnostics platforms, including some of the key testing platforms used to detect the SarsCoV2 virus.
So I bring an insider’s perspective that is scientifically oriented but directed to a general audience trying to make sense of the conflicting stories surrounding the Covid pandemic.
To learn more: https://www.linkedin.com/in/dalewharrison/
If you found this article useful, please sign up by hitting the “Subscribe Now” button below or share it with a friend…





I live in France and there are experts talking about the high mutability of Corona virus all the time.
Armando Arias, virologist at the University of Castilla-La Mancha in Spain and researcher on RNA viruses says:
« RNA viruses (Corona Virus) with smaller genomes, can tolerate higher mutation frequencies (the number of mutations relative to the total number of nucleotides). This number is about 1 mutation per 10,000 nucleotides, which in the world of biology is a lot »
"Large DNA viruses have much lower mutation rates (between 100 and 10,000 times lower). Because their genomes are so large, they can only tolerate one mutation per 10,000 nucleotides. There are many random mutations accumulated in a single genome, which could inactivate some vital functions for the virus. Therefore, DNA viruses are less mutable, "
Esteban Domingo, a virologist at the Severo Ochoa Molecular Biology Center in Madrid says:
« RNA viruses also have polymerases (the enzymes that copy genetic material) that mutate more than DNA viruses. And they have no error repair mechanism.RNA viruses multiply by making mistakes until they end up forming what the scientist calls "Mutants clouds”. »
I’m confused about the virus mutability.
Can you help me to understand it?
Thanks
Eva
Dale- I have found that a low percentage of the articles that I have read even address long Covid. Some that I have seen do refer to “mild” cases that do not require admission to the hospital, yet months on, people are seriously debilitated. Have you seen any statistics on how common this outcome is for mild or asymptomatic Covid infections? Anything on long Covid and contagion?
John Pourchot